Plot composition
Thomas Lin Pedersen
Key takeaway today
Composing plots are no harder than adding a new layer to your plot
Today
- Introduce patchwork for plot composition
- From simple to complex layouts
- Key design principles
- Operators
- Escaping the grid
- The non-panel stuff
- Annotation
- Guide handling
- Object support
- Interactivity
Patchwork
- Composition should have a composable API
- The API should feel natural and easy to reason about in order to invite experimentation
- Assume alignment but give escape hatch
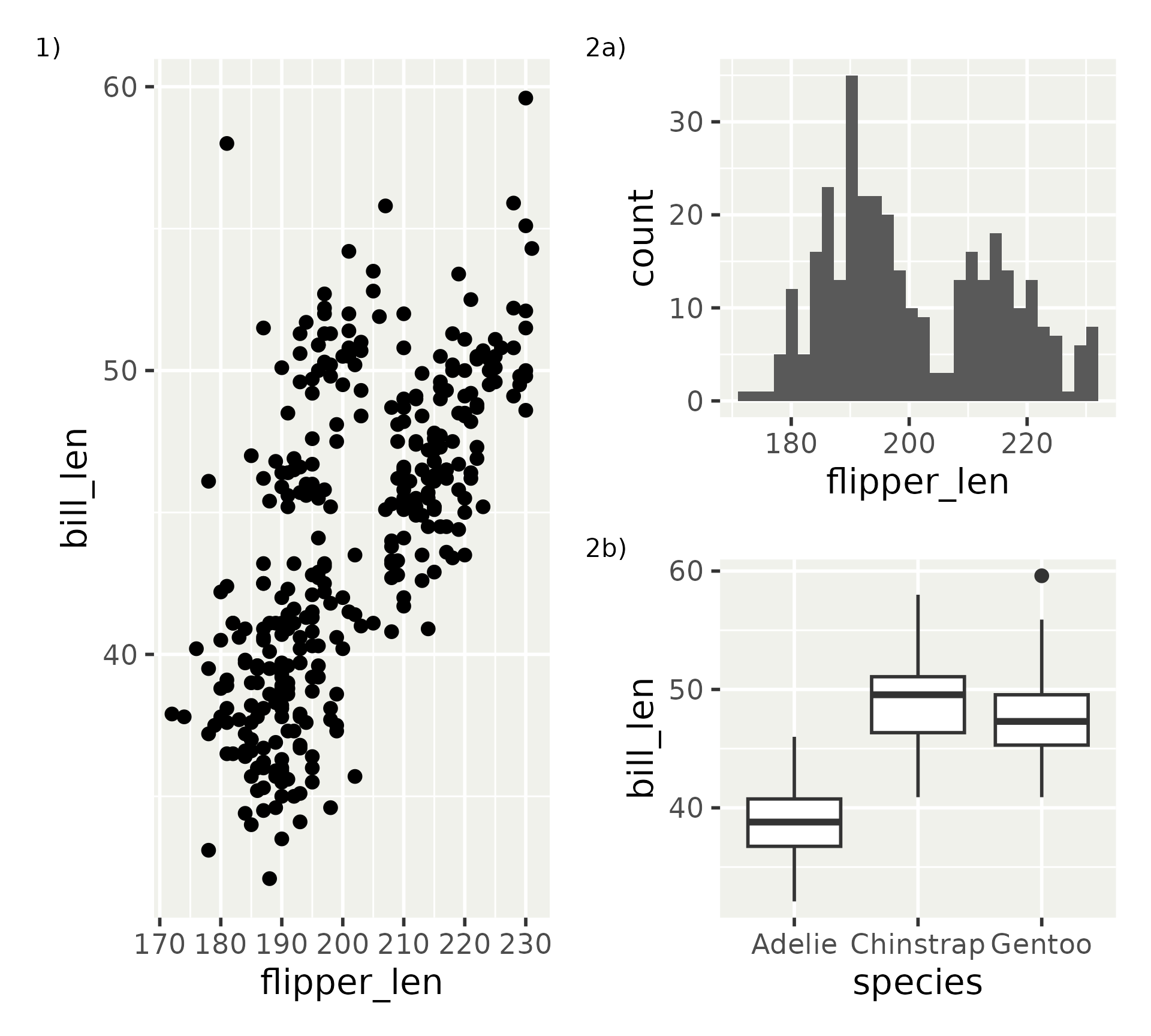
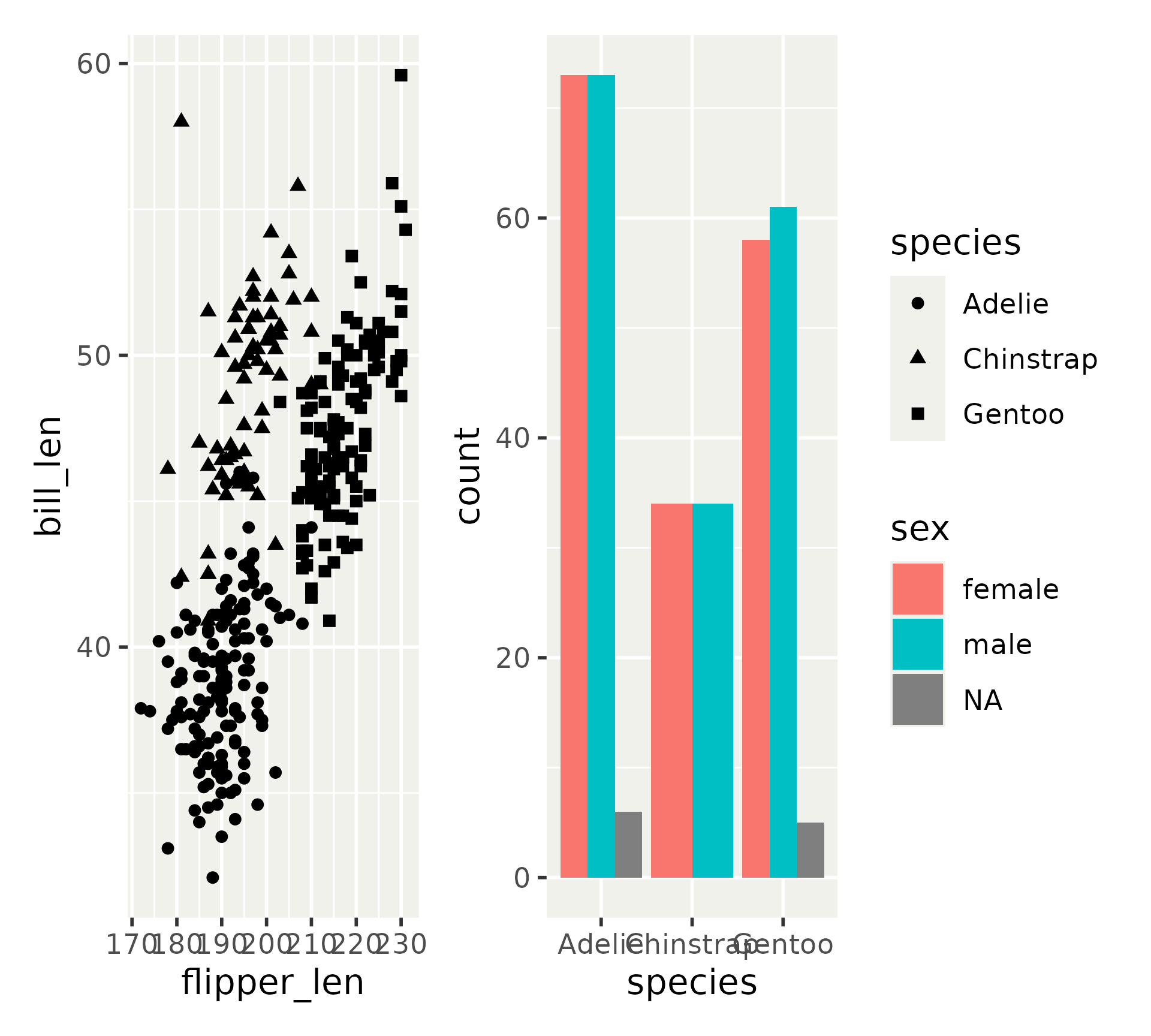
Patchwork in a nutshell
Patchwork in a nutshell
Patchwork in a nutshell
Patchwork in a nutshell
Patchwork in a nutshell
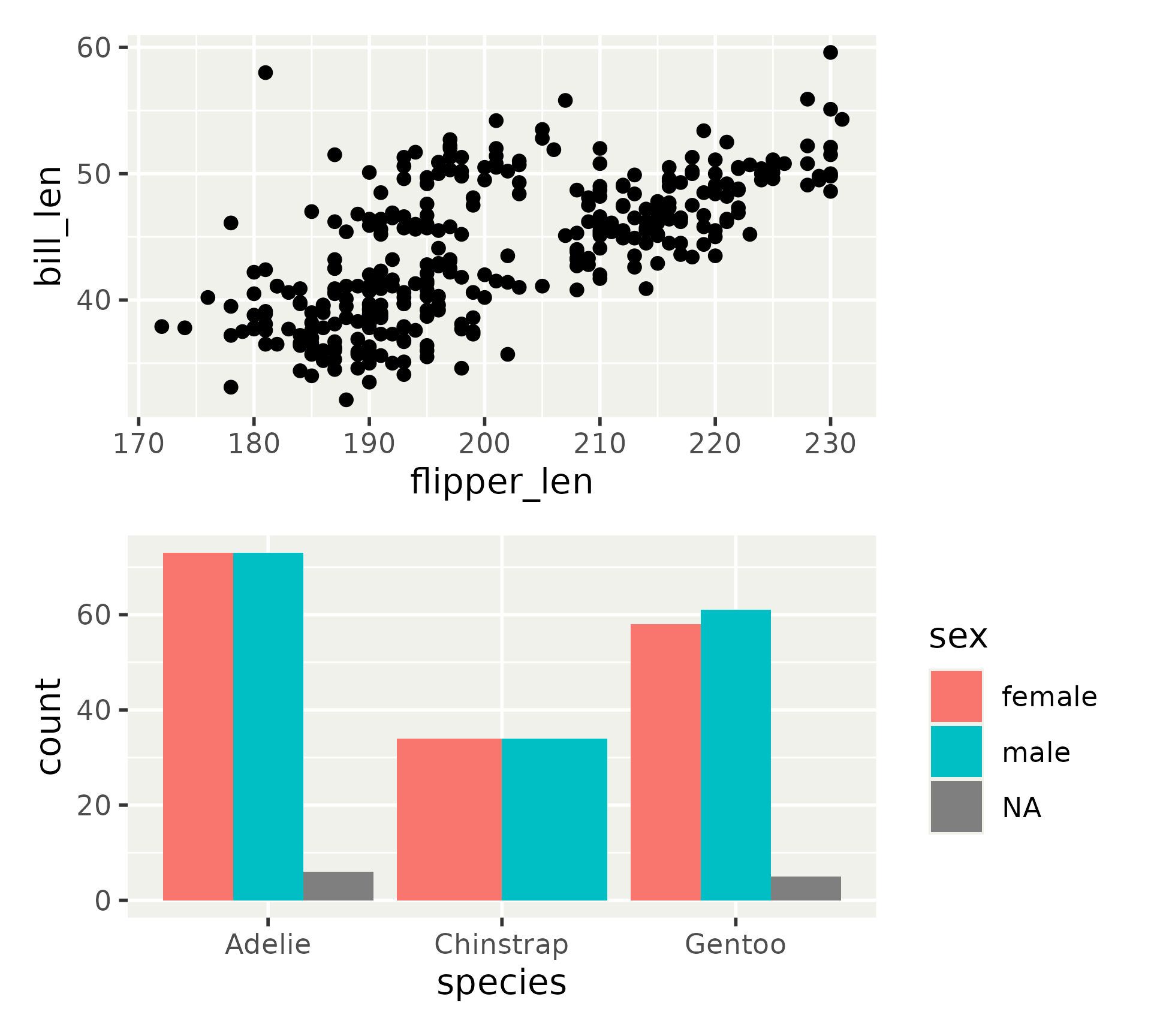
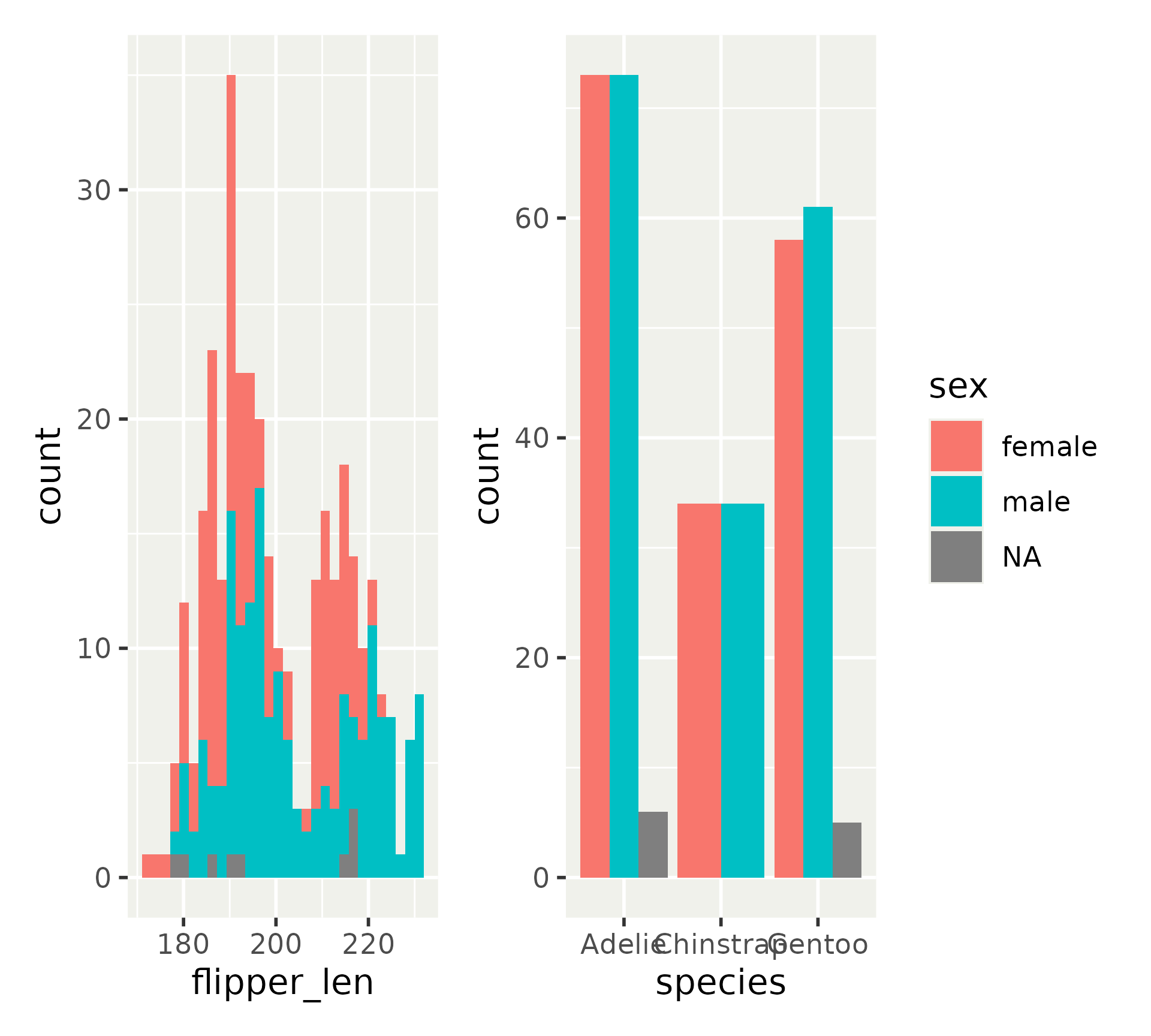
Patchwork layout model

Patchwork layout model

Patchwork layout model

Patchwork layout model

Patchwork layout model

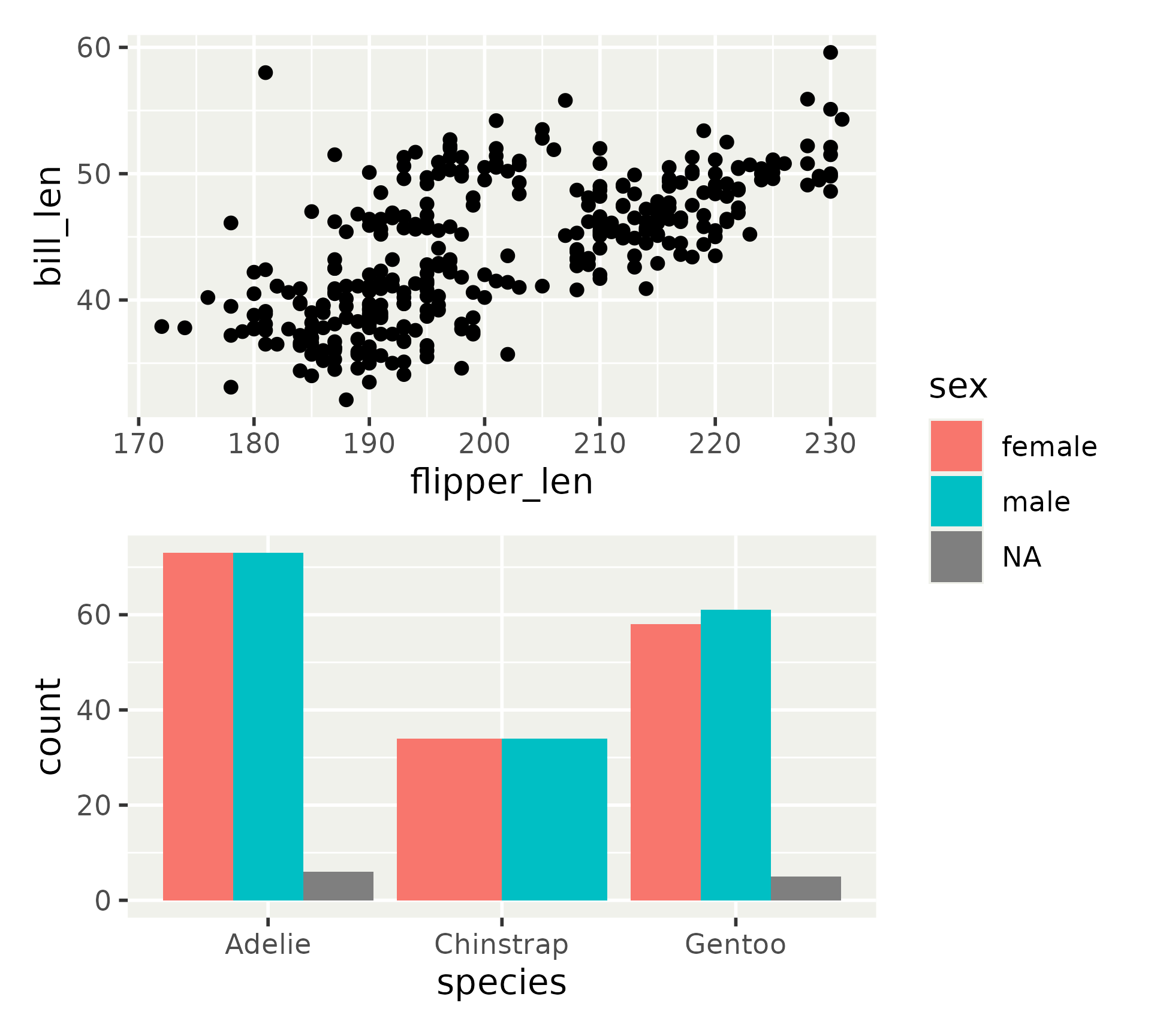
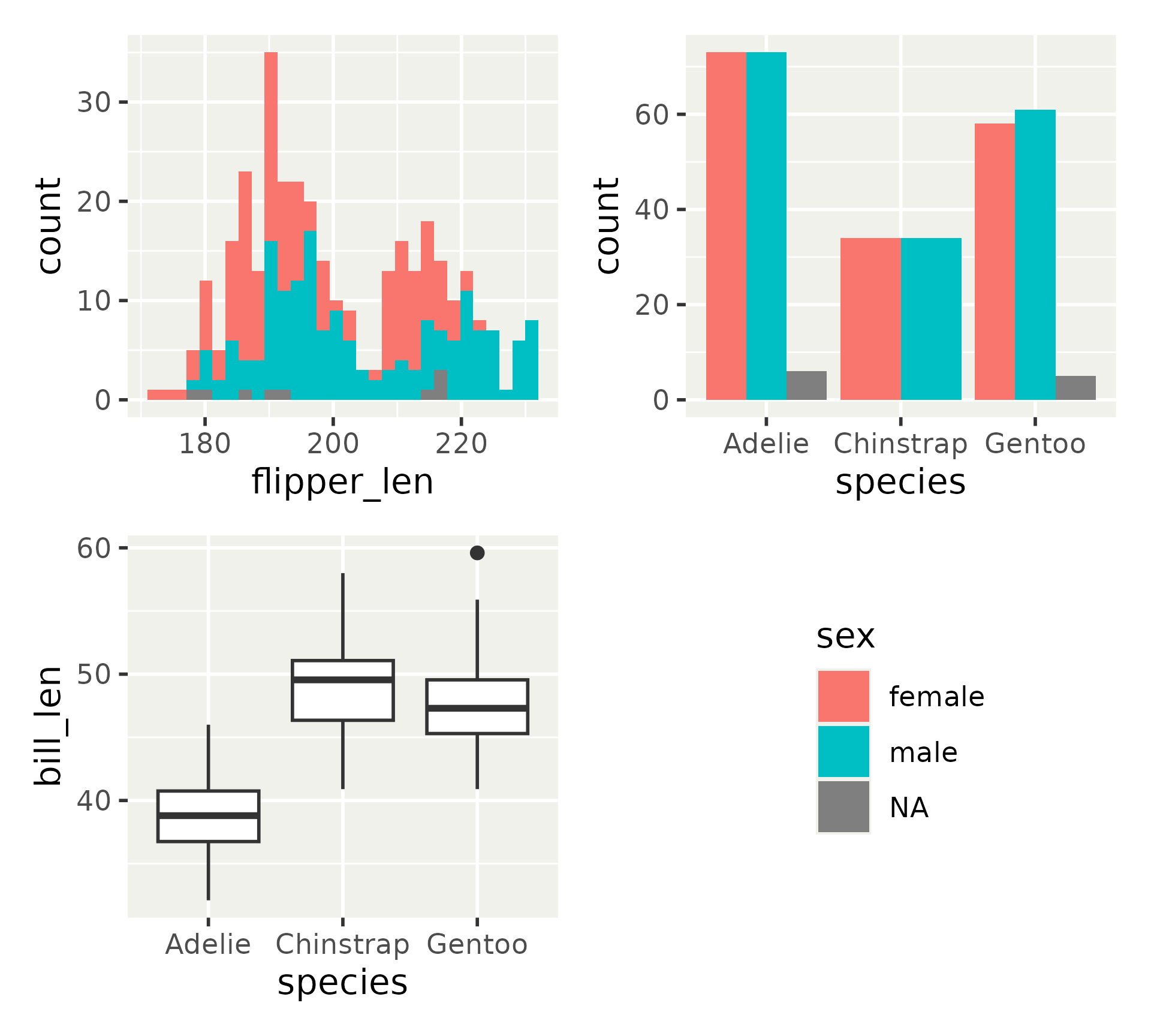
More layout options
More layout options
More layout options
More layout options
Exercise 5.1
05:00
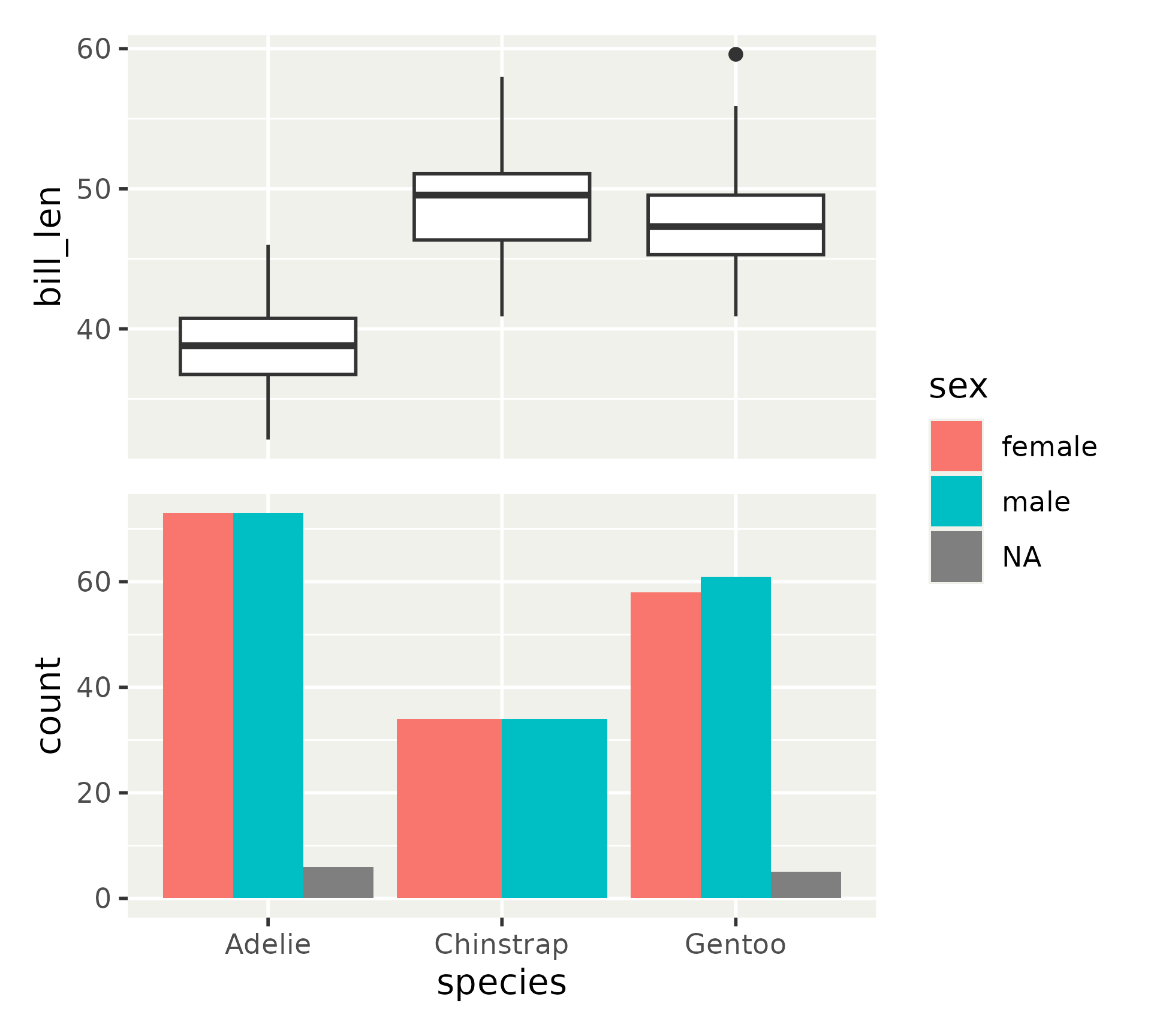
Layout modifiers
The composition consists of a grid
Panels are placed in one or more neighboring cells
Everything outside the panels are placed in the gutter which expands to contain all
What if you don’t want that?
Layout modifiers - Insets
Layout modifiers - Free
Layout modifiers - Free
Layout modifiers - Free
Exercise 5.2
05:00
Annotations
- Once composed, the parts will constitute a new graphic
- Like the individual plots, this graphic can be annotated
Annotations
- Once composed, the parts will constitute a new graphic
- Like the individual plots, this graphic can be annotated
Tagging
With multi-panel graphics you often need a way to refer to the subgraphics
Tagging
It knows about nesting as well
Exercise 5.3
05:00
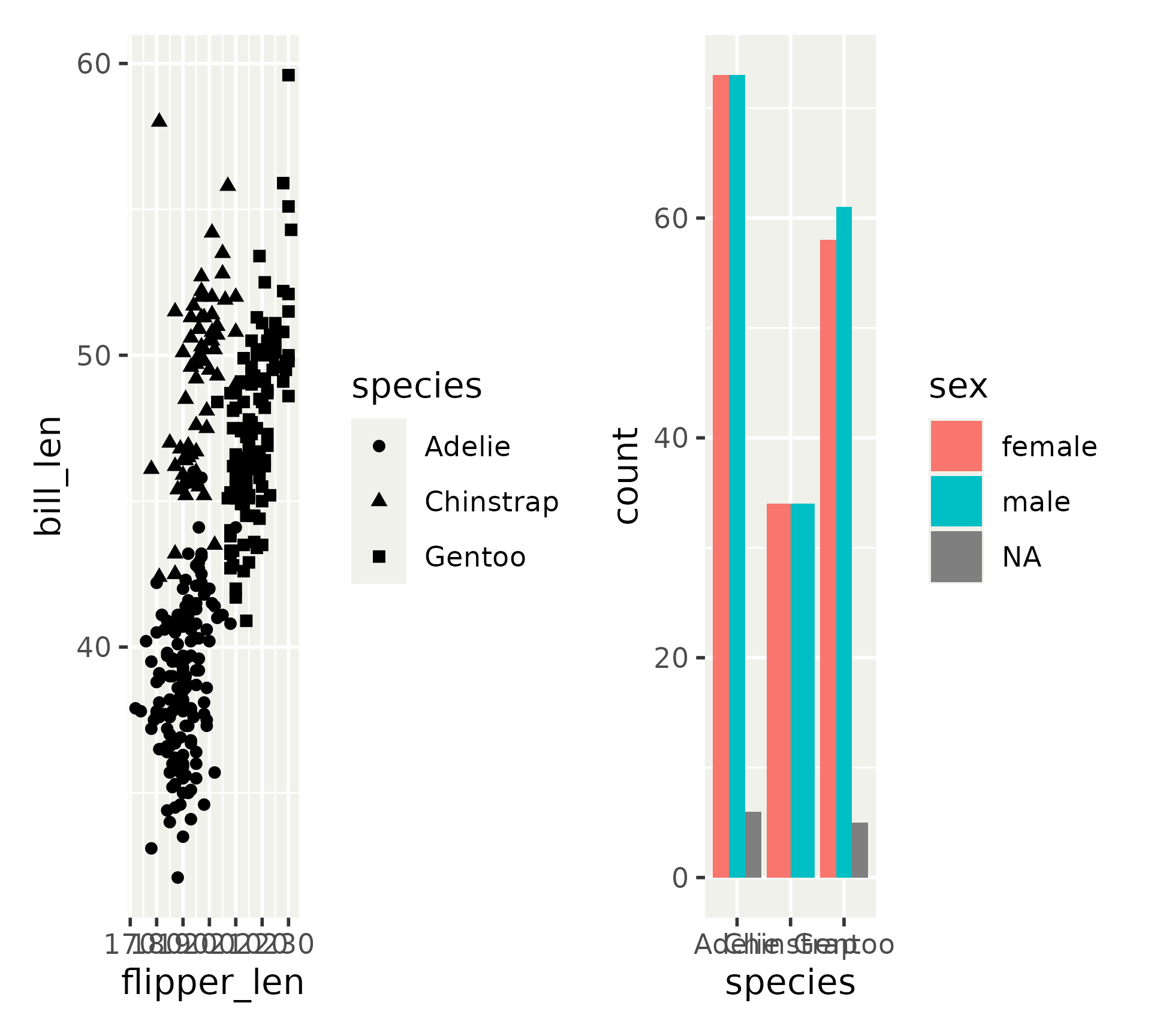
Guide handling
Guides are often global instead of linked to a single subplot
Guide handling
Use guides = "collect" to fetch all guides from subplots and place them at the top level
Guide handling
Guide handling
Guide handling
Duplicate guides are automatically removed
Guide handling
You can place guides inside the composition as well.
Guide handling
Axes are guides too
Exercise 5.4
05:00
Other objects
- Sometimes you need something other than a ggplot (🤯)
- patchwork supports a range of different object types
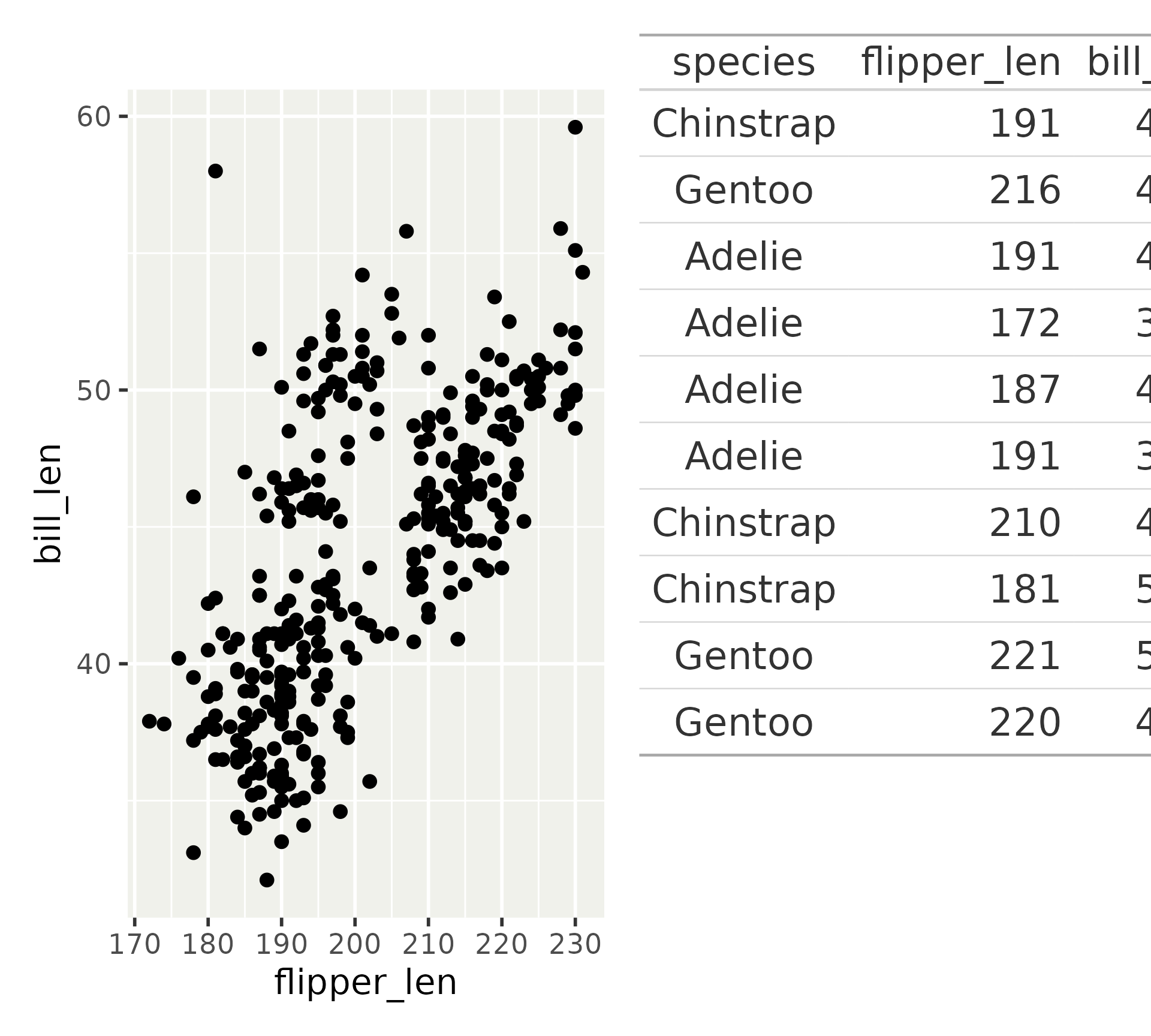
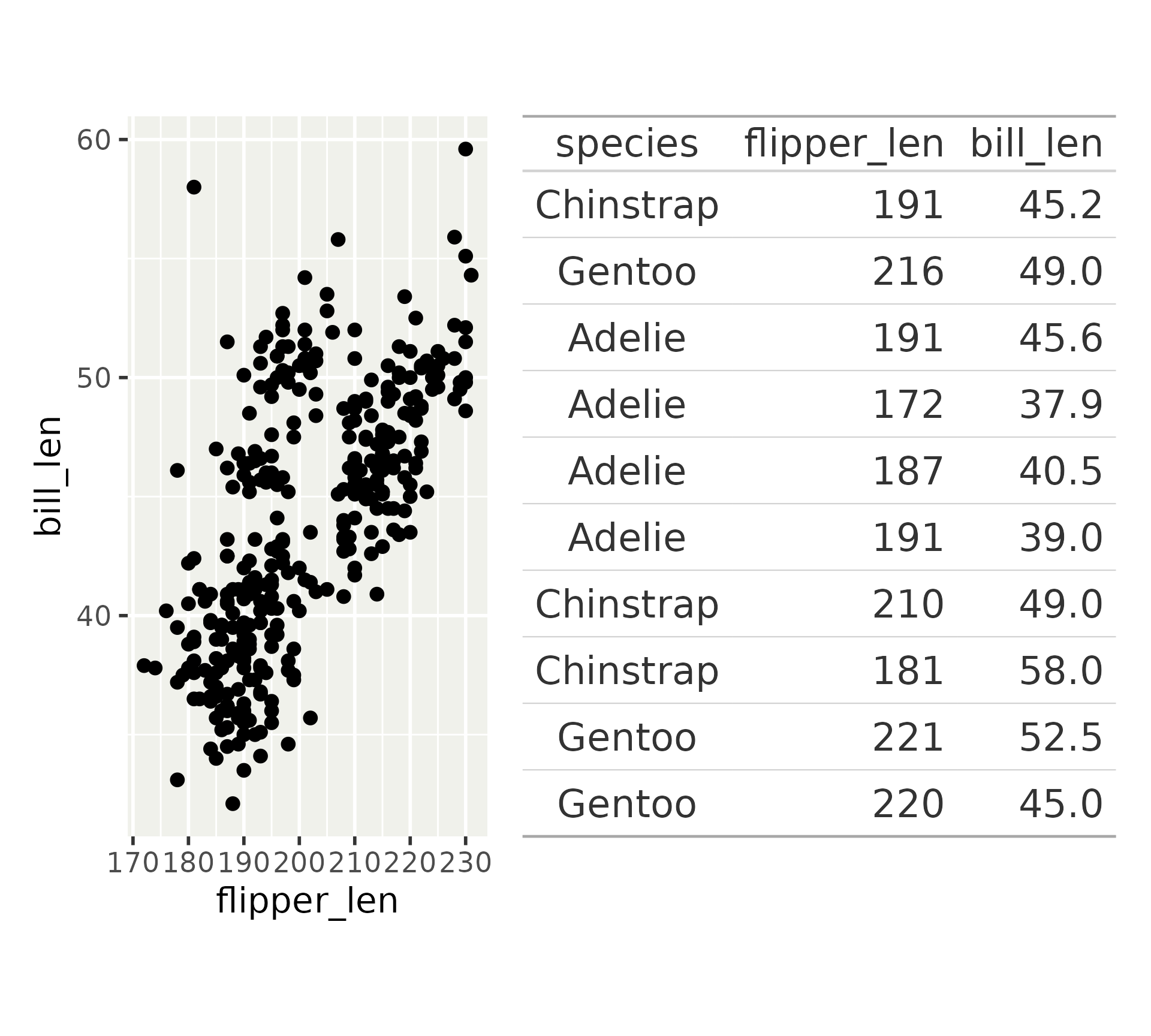
Other objects: gt
Other objects: gt
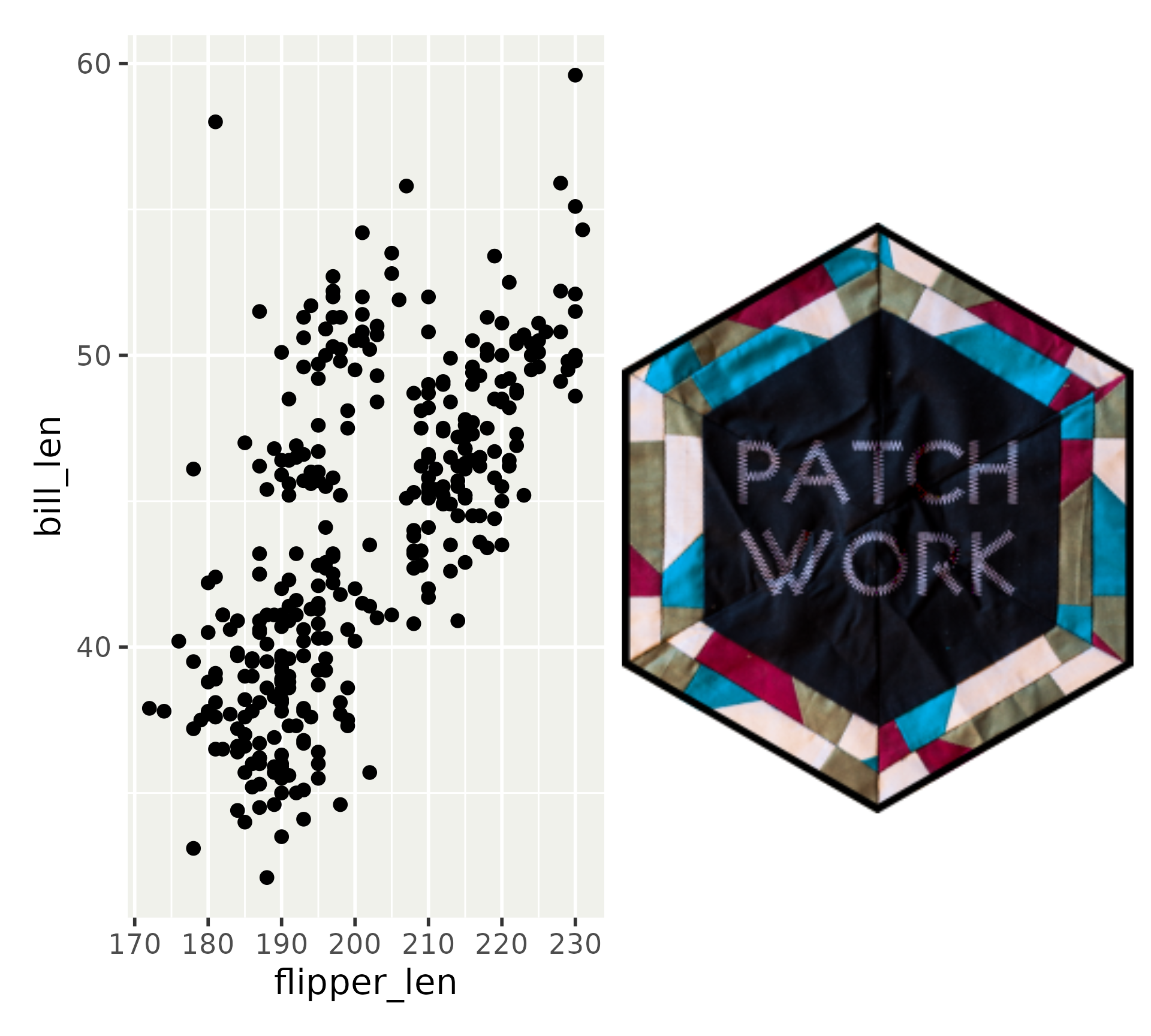
Other objects: images
Other objects: grobs
Other objects: base graphics
Other objects: base graphics
Exercise 5.5
05:00
ggiraph integration
- ggiraph is a package that allows certain interactivity to ggplots
- You create your plots as you always do, but add specific interactive geoms
- If you combine them with patchwork, the interaction is linked
ggiraph integration
ggiraph integration
ggiraph integration
Next session: Creating extensions or Spice up your plot